In this activity, you are going to:
Create a new project from the citizen science dataset and use the clustering feature
Split and concatenate various columns in the dataset
Create a new project from the citizen science dataset and use the clustering feature
1. Let’s create a new project. Start up OpenRefine (if it isn’t running) or click on the OpenRefine logo on the top left to go to the main screen.
Note: If you were working with another project, it has been automatically saved in OpenRefine and the files are stored locally on your computer. You can browse and revisit saved projects by clicking on Open Projects.

2. Click on Create Project. Make sure This Computer is highlighted and then click on Choose Files and browse to the file citizenscience.csv and click on Open. Then click on Next.
3. OpenRefine has recognized the data as CSV. You should now see a preview screen that shows you all the records in the file, nicely laid out with rows and columns. It has some information on users making observations on wildlife in their local area. If that is what you see, keep the defaults, give your project a name at the top and click on Create Project.
4. You should see that there are 1991 rows in our dataset. Click on show 50 rows to show more rows displayed in the window.
5. We have seen text facets before, but we haven't looked at clustering yet. Let's try it out on this dataset. Go to the species guess column (where our citizen scientists have made a guess as to what species they have observed). You may need to scroll to the right to find it. From the species_guess column pull down menu, select Facet > Text facet.
6. You'll see that some of the facets are a bit unusual, and in those cases, you may want to edit them; however, in other cases, you'll see that there are two facets that look very similar, but just have different capitalization, such as "American Pokeweed". When we have facets that look similar, we can use OpenRefine's clustering features to help improve the consistency of the values in that column. Let's take a look. Click on the Cluster button at the top of the facet window.

7. At the top of the new window, you'll see that there are different methods and keying functions you can chose from to find clusters. They roughly go from more strict/unforgiving to looser.
Note: In this case, you should see that the column values are just variations in capitalization, but clustering can also catch typos, plural vs singular and other small differences as well.
8. You can see that it has found entries that it thinks are all referring to the same thing and suggests merging them under one recommended facet. You can put a check mark next to the ones you agree with, and edit the heading that you want to merge them into. Go through and merge the entries found into new terms that have only the first word capitalized by adding a check mark under Merge? and adjusting the New Cell Value.

9. When done, click on Merge Selected & Re-Cluster at the bottom of the window. You might've noticed that as you did a merge, it flashed at the top of the screen how many rows were affected/mass edited.
10. If you no longer have any options with the current method, you could try the nearest neighbour method to get more options, and work through them as well.

As you can see, it is an iterative process to normalize your data. When you're done, with no more options to consider for merging, you can click on Close.
Split and concatenate various columns in the dataset
11. Next let’s manipulate some of our columns. One thing you can do is split values. Let's say we had a column called "names" with values that were in the format "last name, first name". We could use OpenRefine to create two new columns, one for last name and one for first name, and split that names columns into those new columns. Let's see how this works with our example.
We have a column called "scientific_name". With scientific names, the first part is the generic (or genus) name and the second part is the specific name or epithet. So let's split this column so we can see how many of a particular genus were identified. From the scientific_name column pull down menu, select Edit column > Split into several columns.

For the separator, put a space, split into 2 columns at most, and uncheck Remove this column because we want to keep it. Then click on OK.

12. You'll see we now have two new columns to the right of scientific_name: scientific_name 1 & scientific_name 2. From each column’s pull down menu, select Edit column > Rename this column. For the first one, call it "genus". For the second one, call it "epithet".

Now we have our data split into two columns instead of one. If you wanted, you could use text facets on the genus column to see how many of each genus were identified. From the genus column pull down menu, select Facet > Text facet. In the facet window, click on Sort by > count to sort the facets and see which genus names are the most common.


13. Another option is to do the opposite; you can concatenate (join) strings and/or values from two or more columns together. Let's say that we wanted to combine the information on the user id and login into one column with the format "username (user id)". Starting with OpenRefine 3.3, there are now two ways to join columns: a fast method that doesn’t offer much flexibility and a more complex and powerful method.
For this example, we're going to try both methods: we’ll first use OpenRefine’s Join Column option to create a new column to store this information and then use the add column based on this column feature to show some customization options. Depending on what you're doing, sometimes it is better to keep your data intact in its column, and create a new column of data with the changes made, so you can still refer to the original column's values, if need be.
14. First, the simpler method: From the user_login column pull down menu, select Edit Column > Join columns.

15. On the left, drag user_id so that it directly follows user_login, and check it. Change the separator between the content of each column to be a space. Select Write result in a new column named… and name it "User1". Uncheck delete joined columns, and then click OK.

16. There, you have a basic join! Note that there were no options for formatting the text in this new column. To do that, you will need to use GREL.
17. For more advanced formatting options, let’s use the second method.
Note: if you are using OpenRefine version 3.2 or earlier, this more complex method will be your only option.
From the user_id column pull down menu, select Edit column > Add column based on this column.

18. Give the new column the name "User2".
19. Now in order to tell OpenRefine to concatenate values from two columns, we need to use the *General Refine Expression Language (GREL). It is a language that allows you provide more complex instructions to OpenRefine than what you can do using menu options. You can use GREL to perform simple transformations on your data, such as string manipulations.
20. In our starting expression, "value" refers to the value of the current column. If we want to refer to another column in our expression, we use the term cells. and then the name of a column then .value. So for the expression in this case, type:
cells.user_login.value + " (" + value + ")" 
The plus sign is used to join the different values or strings together into one long string. So we're creating a string that is the user login, a space, and then the user id in parentheses.
21. You'll notice that when you type in the expression, the preview at the bottom changes to show you what the resulting value will be. This preview is extremely helpful when writing GREL expressions! If everything looks good, click on OK. Now you should have a new column called User2 made up of information from the "user_id" and "user_login" columns.
*To learn more about the General Refine Expression Language (GREL): https://docs.openrefine.org/manual/expressions#grel-general-refine-expression-language
Restructure the dataset by removing columns and rows, and then work with Undo/Redo to roll those changes back
22. Let's say we're unhappy with entire columns – data we can't do much with, and so we don't want. Well we can remove whole columns. There are a couple ways to do this. One way is from the pull down menu. Let's say we don't want the column tag_list, as it wasn't being consistently used. From the tag_list column pull down menu, select Edit column > Remove this column.

23. Another way to bulk remove columns is to go to the special All column pull down menu on the far left. From the All column pull down menu, select Edit columns > Re-order / remove columns...

From here you can drag columns from the left to the right to remove them – do this for private latitude and private longitude. We can also reorder columns. Move the license, species_guess and quality_grade columns to just under id to move those columns more to the left. Once we're done, click on OK to make the changes.

24. Instead of working with columns, we can also work with rows. Another feature of OpenRefine is the ability to flag or star certain rows and then facet by them. (We will cover faceting by stars and flags later.) Starring and flagging are ways of selecting specific rows that we want to act on later. Although they are functionally the same, in this tutorial we will use flags to select rows we want to delete. An easy way to flag rows is to just click on the flag symbol next to a row of interest – try flagging the first few rows of our dataset.

25. We can also facet our dataset to show certain rows and then automatically flag those rows. For example, to see how many rows have a blank value for a particular column, you can facet by blanks. In this exercise, we will select all rows that both have a blank in the license column and “casual” in their quality grade, and then delete them. From the license column pull down menu, select Facet > Customized facets > Facet by blank (null or empty string)

In the facet box that gets added to the bottom left of your window, click on true to show only the rows where that column is blank (i.e., rows where no license has been specified).

26. Let's cross check that with the ones where they were casual observations. From the quality_grade column pull down menu, select Facet > Text facet. Select casual from the quality_grade facet. Now we have a subset of rows that have a blank for license and are casual observations. Let's say that these 18 rows were no good to us. We could flag them (or star them or remove them). From the All column pull down menu, select Edit rows > Flag rows

27. Finally, reset all facets by clicking on the Reset All button above the facet windows. Now you should see all the rows in your dataset again, some are flagged and some are not.

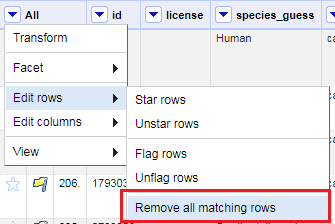
28. Later if you decide that you want to remove those flagged rows that you were unsure of, you can. From the All column pull down menu, select Facet > Facet by flag and then select true from the Flagged Rows facet to show only your flagged rows.

Finally, you can delete all of them. From the All column pull down menu, select Edit rows > Remove all matching rows. All the flagged rows should now be removed from the dataset.
29. Reset all facets again by clicking on the Reset All button above the facet windows to see your remaining rows.
30. We've done a lot to our dataset. But what happens if you do a few things, and then wish you could take some of it back? Well you can with OpenRefine's undo/redo features. Click on the Undo/Redo tab above where the facets show up.

31. You'll see a number of steps that outlines everything you did to this dataset. It is a great way to keep track of what you've done. You can also roll back your changes to a previous version by clicking on the last step you were happy with. Then in the main window, your dataset will look as it did at that point in time. For example, click on the item that says Reorder columns. You'll see that the steps after that have greyed out, which means they haven't happened yet. So for this example, those flagged rows have now not been deleted, and you should see them in your dataset. Be careful: If you go back to a previous step (like we’ve just done), and then start making new changes/transformations - all the subsequent steps will be deleted permanently.

32. Go ahead and try it by starring some rows this time. You should see that the steps we did to flag and delete rows have been replaced by our new starring rows action.
Note: If we had a similarly structured dataset – perhaps for a different snapshot in time – and we wanted to perform the same steps that we had done on this dataset, we could, by clicking on the Extract button.

We would then select the steps we wanted to repeat. You'll see code in the window to the right describing the steps.

You would then copy that code and save it in a text file to keep a copy of your steps. Later if you load up your new dataset, you could go back to the Undo/Redo tab and select Apply and paste in this code into the window to run those steps on the new dataset.

That’s it for our citizen science dataset!
Remember, if you want to now close OpenRefine, first click on the black terminal window and hold CTRL+C until it closes. Then it is safe to close the OpenRefine browser tab.