This tutorial will teach you how to import, clean, explore and visualize public health Covid-19 data using R.
TABLE OF CONTENTS
Importing Data
Data Cleaning
Summary Statistics
Graphs & Maps
Model
Importing Data
You can download Covid-19 data from the John Hopkins Github data. We download the confirmed cases dataset, the deaths dataset and the recovered cases dataset. Note differences in the number of rows and columns.
# IMPORT RAW DATA: Johns Hopkins Github data
confirmedraw <- read.csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv")
str(confirmedraw) # Check latest date at the end of data
deathsraw <- read.csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_deaths_global.csv")
recoveredraw <- read.csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_recovered_global.csv")
Data Cleaning
The next step is the data cleaning process. We want to create country level and global combined datasets with one row per country per date. To do this, we can to convert each dataset from wide format to long format and aggregate at the country level. The cleaned datasets are called confirmed, deaths and recovered. We also combine all three time series datasets into a new time series dataset called country.
# DATA CLEANING library(tidyr) library(dplyr) confirmed <- confirmedraw %>% gather(key="date", value="confirmed", -c(Country.Region, Province.State, Lat, Long)) %>% group_by(Country.Region, date) %>% summarize(confirmed=sum(confirmed)) deaths <- deathsraw %>% gather(key="date", value="deaths", -c(Country.Region, Province.State, Lat, Long)) %>% group_by(Country.Region, date) %>% summarize(deaths=sum(deaths)) recovered <- recoveredraw %>% gather(key="date", value="recovered", -c(Country.Region, Province.State, Lat, Long)) %>% group_by(Country.Region, date) %>% summarize(recovered=sum(recovered)) summary(confirmed)
# Final data: combine all three country <- full_join(confirmed, deaths) %>% full_join(recovered)
We fix the date variable. We create two new variables: the cumulative number of confirmed cases and the number of days variables. We create an aggregate country-level dataset called world.
# Date variable
# Fix date variable and convert from character to date
str(country) # check date character
country$date <- country$date %>% sub("X", "", .) %>% as.Date("%m.%d.%y")
str(country) # check date Date
# Create new variable: number of days
country <- country %>% group_by(Country.Region) %>% mutate(cumconfirmed=cumsum(confirmed), days = date - first(date) + 1)
# Aggregate at world level world <- country %>% group_by(date) %>% summarize(confirmed=sum(confirmed), cumconfirmed=sum(cumconfirmed), deaths=sum(deaths), recovered=sum(recovered)) %>% mutate(days = date - first(date) + 1)
We can extract specific countries from the combined country dataset. We extract Italy here.
# Extract specific country: Italy italy <- country %>% filter(Country.Region=="Italy")
Summary Statistics
We can obtain useful summary statistics from our final cleaned datasets country and world using the summary function and using the by function with the summary function.
# Summary Statistics summary(country) by(country$confirmed, country$Country.Region, summary by(country$cumconfirmed, country$Country.Region, summary) by(country$deaths, country$Country.Region, summary) by(country$recovered, country$Country.Region, summary) summary(world) summary(italy)
Graphs and Maps
# Load graphing package ggplot2 library(ggplot2)
# Barchart of confirmed cases over time - World ggplot(world, aes(x=date, y=confirmed)) + geom_bar(stat="identity", width=0.1) + theme_classic() + labs(title = "Covid-19 Global Confirmed Cases", x= "Date", y= "Daily confirmed cases") + theme(plot.title = element_text(hjust = 0.5))

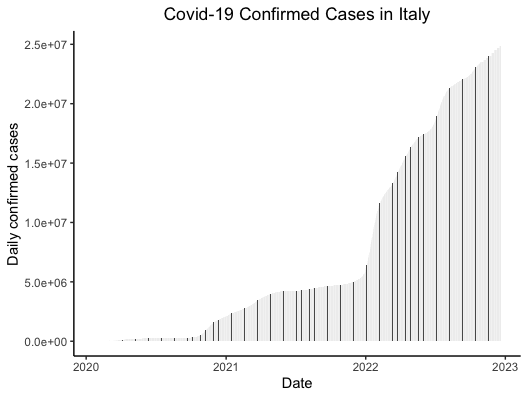
# Barchart of confirmed cases over time - Italy ggplot(italy, aes(x=date, y=confirmed)) + geom_bar(stat="identity", width=0.1) + theme_classic() + labs(title = "Covid-19 Confirmed Cases in Italy", x= "Date", y= "Daily confirmed cases") + theme(plot.title = element_text(hjust = 0.5))
# Line graph of confirmed cases, deaths and recovered - World
str(world)
world %>% select(-cumconfirmed) %>% gather("Type", "Cases", -c(date, days)) %>%
ggplot(aes(x=date, y=Cases, colour=Type)) + geom_bar(stat="identity", width=0.2, fill="white") +
theme_classic() +
labs(title = "Covid-19 Global Cases", x= "Date", y= "Daily cases") +
theme(plot.title = element_text(hjust = 0.5))

# Line graph of confirmed cases over time - World ggplot(world, aes(x=days, y=confirmed)) + geom_line() + theme_classic() + labs(title = "Covid-19 Global Confirmed Cases", x= "Days", y= "Daily confirmed cases") + theme(plot.title = element_text(hjust = 0.5)) # Ignore warning

# Line graph of confirmed cases over time (log10 scale) - World ggplot(world, aes(x=days, y=confirmed)) + geom_line() + theme_classic() + labs(title = "Covid-19 Global Confirmed Cases", x= "Days", y= "Daily confirmed cases (log scale)") + theme(plot.title = element_text(hjust = 0.5)) + scale_y_continuous(trans="log10")

# Line graph of confirmed cases, deaths and recovered - World
str(world)
world %>% select(-cumconfirmed) %>% gather("Type", "Cases", -c(date, days)) %>%
ggplot(aes(x=days, y=Cases, colour=Type)) + geom_line() +
theme_classic() +
labs(title = "Covid-19 Global Cases", x= "Days", y= "Daily cases") +
theme(plot.title = element_text(hjust = 0.5))

# Line graph of confirmed cases on log10 scale for select countries
countryselection <- country %>% filter(Country.Region==c("US", "Italy", "China", "France", "United Kingdom", "Germany"))
ggplot(countryselection, aes(x=days, y=confirmed, colour=Country.Region)) + geom_line(size=1) +
theme_classic() +
labs(title = "Covid-19 Confirmed Cases by Country", x= "Days", y= "Daily confirmed cases (log scale)") +
theme(plot.title = element_text(hjust = 0.5)) +
scale_y_continuous(trans="log10")

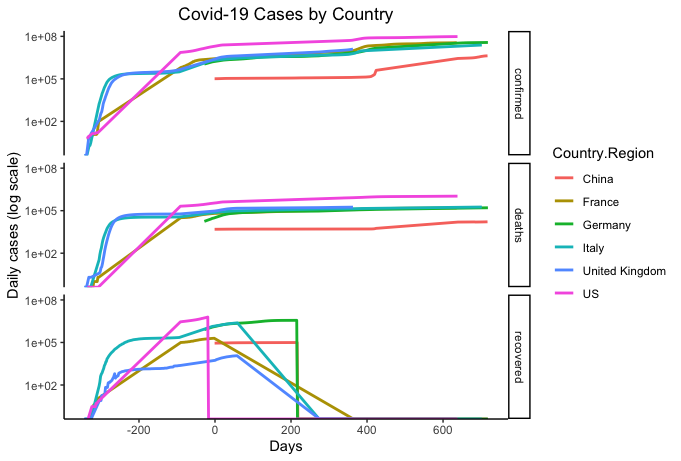
# Matrix of line graphs of confirmed, deaths and recovered for select countries in log10 scale
str(countryselection)
countryselection %>% select(-cumconfirmed) %>% gather("Type", "Cases", -c(date, days, Country.Region)) %>%
ggplot(aes(x=days, y=Cases, colour=Country.Region)) + geom_line(size=1) +
theme_classic() +
labs(title = "Covid-19 Cases by Country", x= "Days", y= "Daily cases (log scale)") +
theme(plot.title = element_text(hjust = 0.5)) +
scale_y_continuous(trans="log10") +
facet_grid(rows=vars(Type))
MAPS
## Map countrytotal <- country %>% group_by(Country.Region) %>% summarize(cumconfirmed=sum(confirmed), cumdeaths=sum(deaths), cumrecovered=sum(recovered))
# Basemap from package tmap library(tmap) data(World) class(World)
# Combine basemap data to covid data
countrytotal$Country.Region[!countrytotal$Country.Region %in% World$name]
list <- which(!countrytotal$Country.Region %in% World$name)
countrytotal$country <- as.character(countrytotal$Country.Region)
countrytotal$country[list] <-
c("Andorra", "Antigua and Barbuda", "Bahrain",
"Barbados", "Bosnia and Herz.", "Myanmar",
"Cape Verde", "Central African Rep.", "", "Congo",
"Dem. Rep. Congo", "Czech Rep.", "Diamond Princess",
"Dominica", "Dominican Rep.", "Eq. Guinea",
"Swaziland", "Grenada", "Holy See", "", "Dem. Rep. Korea",
"Korea", "Lao PDR", "Liechtenstein",
"Maldives", "Malta", "", "Mauritius","",
"Monaco", "MS Zaandam", "", "Macedonia", "",
"Saint Kitts and Nevis", "Saint Lucia", "Saint Vincent and the Grenadines",
"", "San Marino", "Sao Tome and Principe", "Seychelles",
"Singapore", "Solomon Is.", "S. Sudan", "", "Taiwan", "", "",
"United States", "Palestine", "")
countrytotal$Country.Region[!countrytotal$country %in% World$name]
World$country <- World$name
worldmap <- left_join(World, countrytotal, by="country")
worldmap$cumconfirmed[is.na(worldmap$cumconfirmed)] <- 0
# Map
ggplot(data = worldmap) + geom_sf(aes(fill=cumconfirmed), color="black") +
ggtitle("World Map of Confirmed Covid Cases",
subtitle=max(country$date)) +
theme_bw()